

The help section provides information on how to use the FAPS website and API. There are two options for input: Single-Protein Lookup, and a CSV List-Based Search.
With the Single-Protein Lookup, you can search for your protein of interest with an accession ID (e.g. "P08670", "Q9Y490"). You can also upload a .csv file with a column named 'Accession' in the CSV List-Based Search option to filter by. The output will return the simulated 3-D structure of proteins and some experimentally determined secondary structure information. This includes the overall alpha, beta, random coil, and unknown constitute calcuated with both simulated and experimental data for each protein.
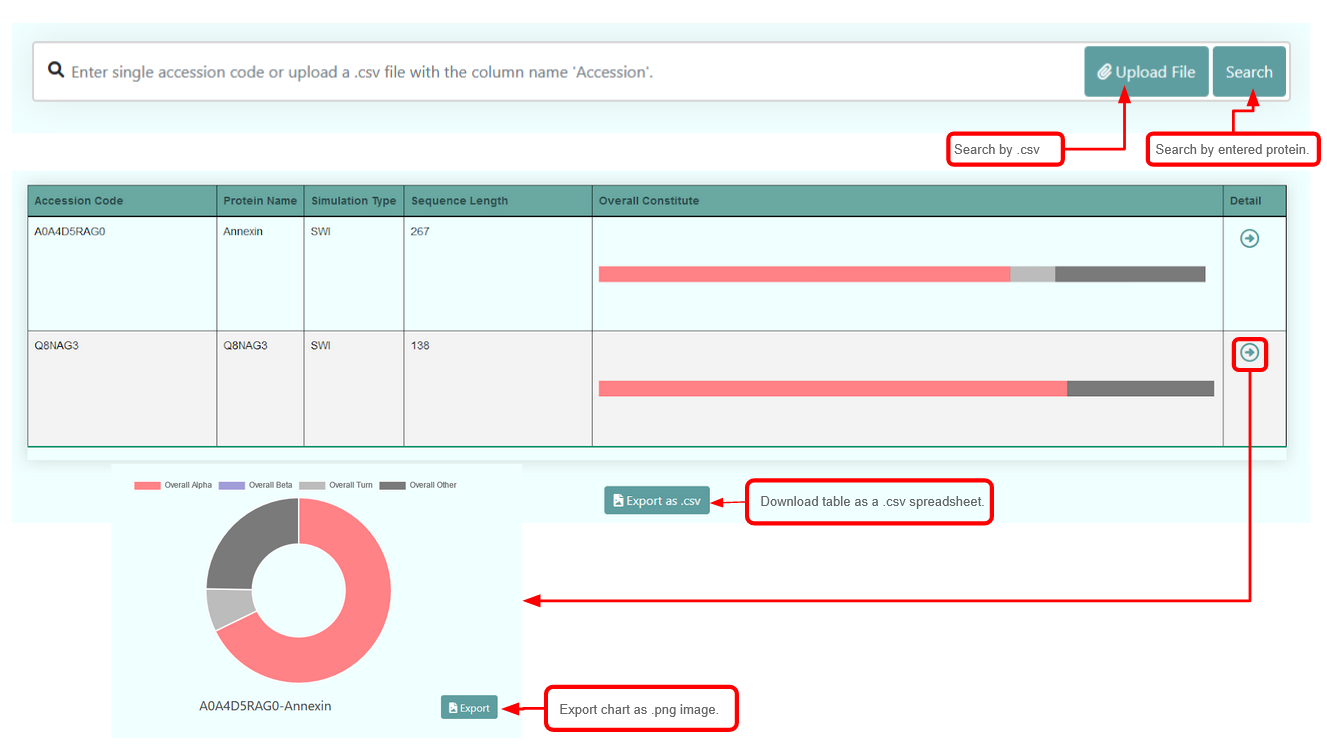
The result summarizes the set of secondary structure information from the protein of interest in a table, and also in a pie chart. The first column of the table indicates the accession ID/protein ID that was searched for; the second column provides information about the simulation type the data was generated from; the third column is the overall amino acid sequence length of the protein; the fourth column presents a bar chart summary of the protein's overall secondary structure. The final column allows you to view a more detailed summary of the results, along with the aforementioned pie chart. The data table can also be exported as a .csv file for raw proportion data, and the pie chart can be exported as an image file.
For each protein, the proportion of simulated secondary structure present was calculated
by dividing the length of each structure type found in the protein by the overall
amino acid sequence length. The experimentally determined data was scraped from UniprotKB and
converted to a proportional format to match the simulated data. The overall constitute for each
was then calculated as the sum total of the product of the simulated proportion and known proportion with
the product of the UniprotKB proportion with the unknown.